Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
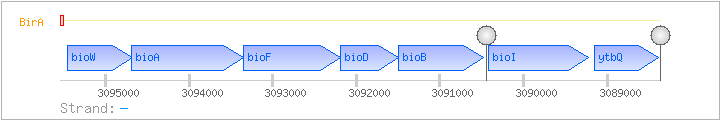
| Regulated Operon: | bioWAFDBI-ytbQ |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| bioW | - | 3094679..3095455 | COG1424H | |||
| bioA | - | 3093343..3094689 | COG0161H | x0337-BAC | ||
| bioF | - | 3092184..3093353 | COG0156H | bioF-BAC | ||
| bioD | - | 3091492..3092187 | COG0132H | |||
| bioB | - | 3090482..3091489 | COG0502H | |||
| bioI | - | 3089226..3090413 | COG2124Q | |||
| ytbQ | - | 3088388..3089149 | COG0451MG |
| Operon evidence: | Northern blotting (7.0 kb transcript found by Bower et al.; 7.2 kb transcript found by Perkins et al.). |
|---|---|
| Reference: | Bower S, et al. (1996), Perkins JB, et al. (1996), Genbank AF008220 |
| Comments: | The readthrough terminator downstream of bioB leads to a 5.0 kb (as measured by Bower et al.) / 5.1 kb (as measured by Perkins et al.) bioWAFDB transcript. Readthrough termination was confirmed by Perkins et al. by deleting the stem-loop structure downstream of bioB. Perkins et al. also found a monocistronic bioW transcript of 0.8 kb, which may be an RNA degradation product. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| BirA | Negative | ND | 3095505..3095544 | CTAATTGTTAACCTTTGAATATAATTGGTTAACAATTTAG |
Bower S, et al. (1996): DB Perkins JB, et al. (1996): DB NB |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| GAACGGAGCATAATCATTTTCTAAGATTATGCTCTTTTTCTTTTGTTAT >>>>>>>>>> <<<<<<<<<< |
ytbQ | |||
| AAAAGCAATCGGTATGATGTCGATTGTTTTTATTTTTGAAC >>>>>>>> <<<<<<<< |
bioB |
|