Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
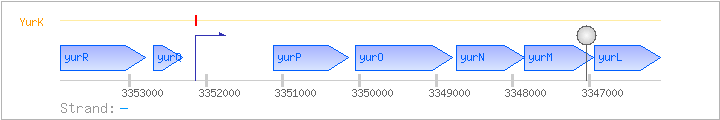
| Regulated Operon: | yurRQPONML |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| yurR | - | 3352789..3353907 | COG0665E | x0835-BAC | ||
| yurQ | - | 3352312..3352686 | COG0322L | |||
| yurP | frlB | - | 3350137..3351123 | |||
| yurO | frlO | - | 3348788..3350056 | |||
| yurN | frlN | - | 3347852..3348730 | |||
| yurM | - | 3346946..3347848 | ||||
| yurL | - | 3346078..3346932 |
| Operon evidence: | Northern blotting (7.5 kb transcript) |
|---|---|
| Reference: | Yoshida K, et al. (2003) |
| Comments: | The long mRNA molecule agrees poorly with the sequence and with the microarray data. The Northern blotting results also showed yurP (1.0 kb) and yurPO (2.5 kb) transcripts. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| YurK | Negative | +1:+50 | 3352094..3352143 | ATATAATGTTATATAACGTTATATAATAAAAACGAGGAGTGAAGGATTTG |
Deppe VM, et al. (2011): PE GS RG |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AAAAACGAGAATACTATAAGCAGTGTCTCGTTTTTTATGCCTGTT >>>>>>>>>> <<<<<<<<< |
yurM |
|