Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
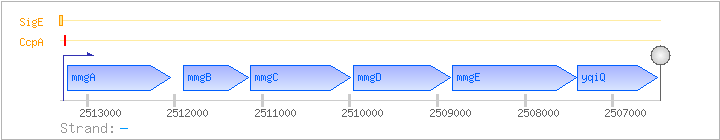
| Regulated Operon: | mmgABCDE-yqiQ |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
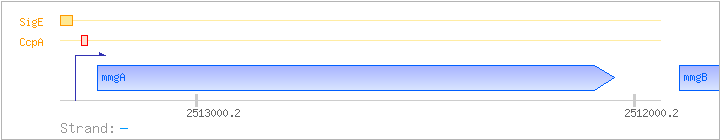
| mmgA | yqiL | - | 2512046..2513227 | acetyl-CoA acetyltransferase | COG0183 | mmgA-BAC |
| mmgB | yqiM | - | 2511162..2511896 | 3-hydroxybutyryl-CoA dehydrogenase | COG1250 | |
| mmgC | yqiN | - | 2509998..2511134 | acyl-CoA dehydrogenase | COG1960 | x0817-BAC x0941-BAC |
| mmgD | yqiO | - | 2508846..2509964 | citrate synthase III | COG0372 | mmgD-BAC |
| mmgE | yqiP | - | 2507413..2508831 | COG2079 | mmgE-BAC-1 mmgE-BAC-2 | |
| yqiQ | - | 2506490..2507395 | COG2513 | yqiQ-BAC |
| Operon evidence: | lacZ transcriptional fusion; Northern blotting |
|---|---|
| Reference: | Bryan EM, et al. (1996), Eichenberger P, et al. (2003), BSORF |
| Comments: | The transcriptional fusion shows that the operon includes mmgE, but does not exclude the possibility that genes further downstream are also part of the operon. The Northern blotting results in BSORF show a mmgABCDE-yqiQ transcript, which agrees with the predicted transcriptional terminators. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| CcpA | Negative | +15:+28 | 2513250..2513263 | TGTAAGCGCTGTCT |
Bryan EM, et al. (1996): SDM DB |
| SigE | Promoter | -39:+4 | 2513274..2513316 | TCATTCATTCATGCCCGTTTCAAAGCATACATTCATAGAAGAC |
Bryan EM, et al. (1996): RG OV PE |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AAAAAGCCGGAGCATTCTCCGGCTTTTTTTCAGCTATC >>>>>>> <<<<<<< |
yqiQ |
|