Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
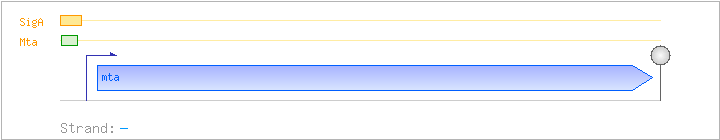
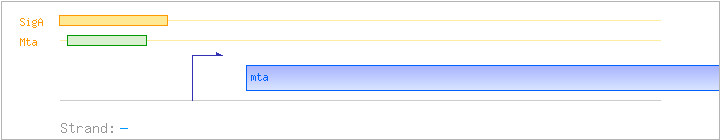
| Regulated Operon: | mta |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
| mta | ywnD | - | 3763155..3763928 | transcriptional regulator (MerR family) | COG0789 | mta-BAC |
| Operon evidence: | upstream and downstream gene are on the opposite strand |
|---|---|
| Reference: | Presecan E, et al. (1997), Genbank Y08559 |
| Comments: |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| Mta | Positive | -35:-13 | 3763956..3763978 | GACCCTAACGTTGCGTGATTGTT |
Baranova NN, et al. (1999): NB FT |
| SigA | Promoter | -41:+4 | 3763940..3763984 | GGGATTGACCCTAACGTTGCGTGATTGTTTACGATAAAAAAGAGG |
Baranova NN, et al. (1999): RO |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| GAAAACCCCCGGCCGTAAAGGCCGGGGGTTTACGCATCTTATA >>>>>>>>> <<<<<<<<< |
mta |
|