Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
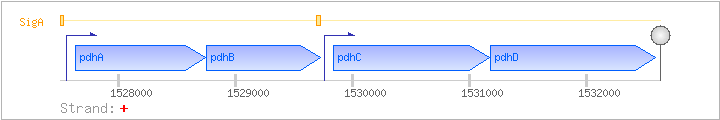
| Regulated Operon: | pdhABCD |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
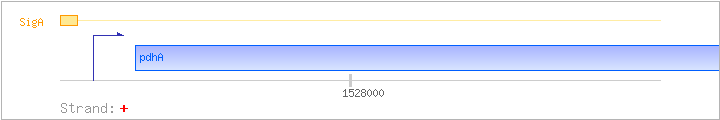
| pdhA | aceA | + | 1527633..1528748 | pyruvate dehydrogenase (E1 alpha subunit) | COG1071 | pdhA-BAC pdhA-STA aceA-BAC |
| pdhB | + | 1528752..1529729 | pyruvate dehydrogenase (E1 beta subunit) | COG0022 | ||
| pdhC | + | 1529844..1531172 | pyruvate dehydrogenase (dihydrolipoamide acetyltransferase E2 subunit) | COG0508 | pdhC-BAC | |
| pdhD | citL | + | 1531177..1532589 | dihydrolipoamide dehydrogenase E3 subunit of both pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase complexes | COG1249 |
| Operon evidence: | Northern blotting (5.2 kb transcript) |
|---|---|
| Reference: | Hemila H, et al. (1990), Gao H, et al. (2002) |
| Comments: | Hemila also found a 2.8 kb pdhCD and a 1.7 kb monocistronic transcript. However, Gao et al. note that promoters were found in front of pdhA and pdhC only; no promoter was found in front of pdhD. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigA | Promoter | -64:+12 | 1527498..1527573 | AAGGTTTTGGCAAACTGGGGATCGGCTAAAATATAATACGACTTACTGCTGATACTTTAGGGTACCCTAAGTTTGT |
Gao H, et al. (2002): NB PE Hemila H, et al. (1990): NB |
| SigA | Promoter | -70:+8 | 1529690..1529767 | AGACGTTCTTGAAACAGCAAGAAAAGTGCTTGAATTTTAATCAAACTGCATAATCGAGAGGGAAGATGAACGTTTTCC |
Gao H, et al. (2002): NB PE Hemila H, et al. (1990): NB |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AAAAACAGCCCCGCTTTGAGCGAGGGCTGTTTTTTTATTTTGAC >>>>>>>>>> <<<<<<<<<<< |
pdhD |
|