Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
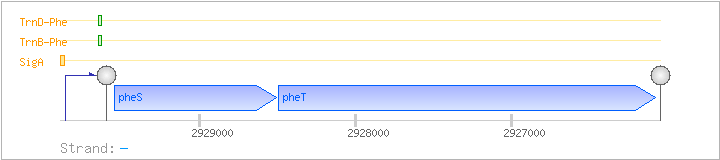
| Regulated Operon: | pheST |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
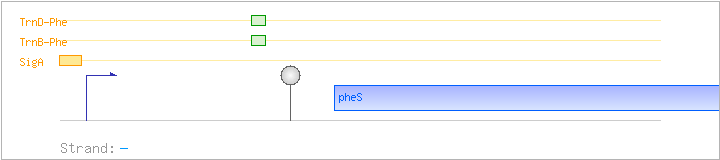
| pheS | - | 2928506..2929540 | phenylalanyl-tRNA synthetase (alpha subunit) | COG0016 | pheS-BAC pheS-LAB pheS-STA pheS-STR | |
| pheT | - | 2926076..2928490 | phenylalanyl-tRNA synthetase (beta subunit) | COG0072 | pheS-LAB pheT-BAC pheT-CLO |
| Operon evidence: | Genome analysis |
|---|---|
| Reference: | Brakhage AA, et al. (1990), Grundy FJ & Henkin TM (1993) |
| Comments: | Transcripts starting about 1.0 kb upstream of the indicated promoter were also detected. These may correspond to a ysgA-pheST transcript. Based on homology, transcriptional readthrough is expected to occur at the stem-loop upstream of pheS if uncharged phenylalanyl tRNA binds to the T-box motif in the nascent mRNA transcript. The tRNA anticodon binds to the UUC codon in the sequence GCCUUCACC in the pheS leader mRNA. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigA | Promoter | -38:+5 | 2929854..2929896 | AGTGTTGCGCCAAGCGTTGGATTTCTCTATAATAGAACATAAT |
Brakhage AA, et al. (1990): PE S1 |
| TrnB-Phe | Positive | ND | 2929619..2929657 | AGTCTGTCTGAAATAAGGGTGGTACCGCGGCCACAACTC |
Grundy FG & Henkin TM (1993): HM |
| TrnD-Phe | Positive | ND | 2929619..2929657 | AGTCTGTCTGAAATAAGGGTGGTACCGCGGCCACAACTC |
Grundy FG & Henkin TM (1993): HM |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| TACCGCGGCCACAACTCGTCCCTTGTACAAGGGACGGGTTTTTTTTATTTTCC >>>>>>>>>> <<<<<<<<<< |
pheS | |||
| TTAAGAGGCTGATAAAAAAGCTTGCAGAGAAATCTGCAAGCTTTTTTCTATGAACG >>>>>>>>> <<<<<<<<< |
pheT |
|