Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
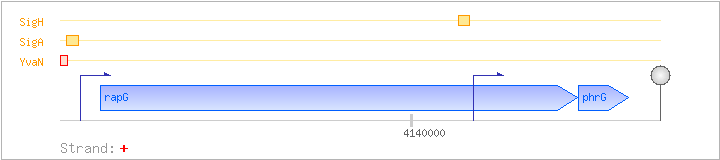
| Regulated Operon: | rapG-phrG |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
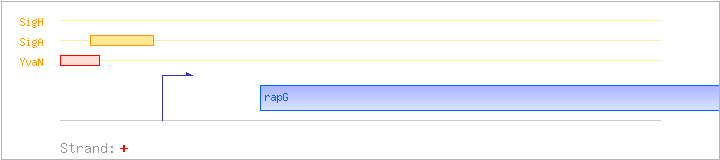
| rapG | yycM | + | 4139285..4140382 | response regulator aspartate phosphatase | COG0457 | |
| phrG | yycL | + | 4140383..4140499 | regulator of the activity of phosphatase RapG |
| Operon evidence: | Northern blotting (1.6 kb transcript); primer extension of phrG |
|---|---|
| Reference: | McQuade RS, et al. (2001), Ogura M, et al. (2003), BSORF |
| Comments: | Internal promoter in front of phrG leads to monocistronic phrG transcripts of 0.6 kb and 0.4 kb. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| YvaN | Negative | -55:-27 | 4139185..4139213 | CTTCATATATTGACTAGAGGATTTTACAT |
Hayashi K, et al. (2006): RG DB AR PE GS FT |
| SigA | Promoter | -34:+5 | 4139206..4139244 | TTTTACATAAAATAGAAAGAGGTGTTACTATCAGAATAA |
Hayashi K, et al. (2006): PE |
| SigH | Promoter | -38:+4 | 4140105..4140146 | TGAAGGAAAAATTGACTGCTGAGCCGAATAGAATATGTGAGG |
McQuade RS, et al. (2001): PE RG DB Ogura M, et al. (2003): DB NB RG |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AAGAATCCTAAAACGGTTTGTAGTTTTAGGATTCTTTCATCTTTTC >>>>>>>>> <<<<<<<<< |
phrG |
|