Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
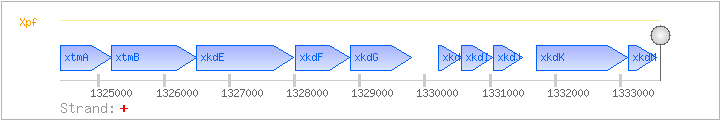
| Regulated Operon: | xtmAB-xkdEFGHIJKM |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
| xtmA | ykxF | + | 1324404..1325201 | PBSX defective prophage terminase (small subunit) | COG3728 | |
| xtmB | ykxG | + | 1325198..1326499 | PBSX defective prophage terminase (large subunit) | COG1783 | |
| xkdE | + | 1326503..1327990 | ||||
| xkdF | + | 1328010..1328837 | ||||
| xkdG | + | 1328863..1329798 | xkdG-BAC | |||
| xkdH | + | 1330202..1330558 | ||||
| xkdI | + | 1330555..1331040 | ||||
| xkdJ | + | 1331053..1331493 | ||||
| xkdK | + | 1331712..1333106 | ||||
| xkdM | + | 1333113..1333556 |
| Operon evidence: | Genome analysis |
|---|---|
| Reference: | Wood HE, et al. (1990) |
| Comments: | Wood et al. assume an operon of at least 19 kb, presumably including the downstream genes xkdNOPQRSTUVWX-xepA-xhlAB-xlyA |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| Xpf | Positive | ND | ND | ND |
McDonnell GE, et al. (1994): DB |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| ATACGCAGACCTTTCTCAGAAAGGTCTGTTTTTTAAAGATGAA >>>>>>>>>> <<<<<<<<<< |
xkdM |
|