Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
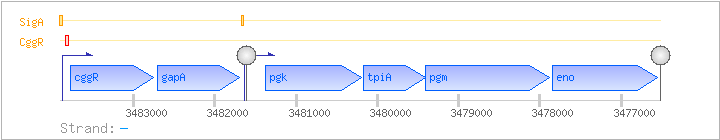
| Regulated Operon: | cggR-gapA-pgk-tpiA-pgm-eno |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| cggR | yvbQ | - | 3482752..3483774 | COG2390K | ||
| gapA | gap | - | 3481698..3482705 | COG0057G | gapA-BAC-1 gapA-MYC-1 gapA-BAC-1 gapA-MYC-1 gapA-MYC-2 stu1788-STA stu1788-STR | |
| pgk | gerG | - | 3480197..3481381 | COG0126G | pgk-BAC pgk-CLO pgk-LAB pgk-STA | |
| tpiA | tim | - | 3479405..3480166 | COG0149G | tpi-BAC tpi-LAB tpi-STA tpi-STR | |
| pgm | - | 3477877..3479412 | COG0696G | x0574-STR | ||
| eno | - | 3476555..3477847 | COG0148G | eno-BAC eno-MYC eno-STA eno-STR |
| Operon evidence: | Northern blotting (7.4 kb transcript) |
|---|---|
| Reference: | Ludwig H, et al. (2001), Genbank L29475 |
| Comments: | The readthrough terminator downstream of gapA leads to a 2.2 kb cggR-gapA transcript and a 6.2 kb gapA-pgk-tpiA-pgm-eno transcript. The internal promoters in front of gapA and pgk lead to a 1.2 kb monocistronic gapA transcript and a 4.8 kb pgk-tpiA-pgm-eno transcript. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| CggR | Negative | +27:+72 | 3483799..3483844 | CTATGACGGGACGTTTTTTGTCATAGCGGGACATATAATGTCCAGC |
Kodama T, et al. (2000): PE RG Doan T, et al. (2003): DB FT |
| SigA | Promoter | -42:+14 | 3483857..3483912 | CTTAACAGTTGAATAAACAATTCCACCCTGTTAAAATAATTAAAGAAAGCAGAAAT |
Ludwig H, et al. (2001): PE |
| SigA | Promoter | -49:+9 | 3481628..3481685 | AAAGGACCTGACTTGGTTCTTTCGAATAGAAGCGCTATAATGAAAGCGGACAAGGGAA |
Ludwig H, et al. (2001): PE |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AAAGACCGCCGGGATTTTCTCTCGGCGGCTTTTTTATGCTTTCA >>>>>>>>> <<<<<<<<< |
eno | |||
| GACAAGGGAAGGGGACGGACTCCCTTTCCCTTTTTCCATGAAGAC >>>>>>>>>> <<<<<<<<<< |
gapA |

|