Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
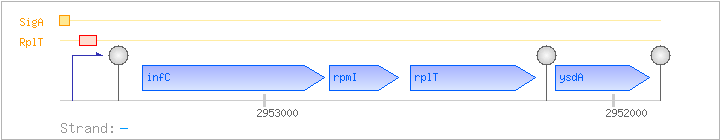
| Regulated Operon: | infC-rpmI-rplT-ysdA |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| infC | - | 2952828..2953349 | COG0290J | infC-BAC infC-STA infC-STR | ||
| rpmI | - | 2952615..2952815 | COG0291J | rpmI-BAC rpmI-STA rpmI-STR | ||
| rplT | - | 2952224..2952583 | COG0292J | rplT-MYC rplT-BAC rplT-LAB rplT-MYC rplT-STA rplT-STR | ||
| ysdA | - | 2951898..2952167 | COG3326S |
| Operon evidence: | Northern blotting (1.7 kb transcript) |
|---|---|
| Reference: | Wipat A, et al. (1996), Choonee N, et al. (2007) |
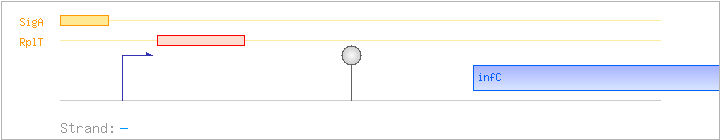
| Comments: | The full-length infC-rpmI-rplT-ysdA transcript is minor; the major transcript has a length of 1.4 kb and includes infC-rpmI-rplT, terminating at the terminator downstream of rplT. Transcriptional readthrough at this terminator was confirmed by RT-PCR experiments. The stem-loop upstream of infC is part of a terminator/antiterminator structure, as shown by Northern blotting, point mutations, lacZ fusions, and in-vitro transcription assays. Binding of the ribosomal protein L20 (RplT) to the nascent mRNA causes transcriptional termination. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| RplT | Negative | +19:+73 | 2953478..2953532 | AAGCAGAAGCACCCGCTTCTCACCTGATTGACACATGCATGTAGTTGGCAGGTTG |
Choonee N, et al. (2007): NB RG SDM, in-vitro assay, OV FT, toeprint |
| SigA | Promoter | -37:+2 | 2953549..2953587 | CTTGACTAAAGATCCGGTATTGTGTAGAATAGTTATTGA |
Choonee N, et al. (2007): PE |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| ATGTTCGTAGTGTGGGTGTTTTATAATGCCCACACTTTTTGTTTGCCTGC >>>>>>>>>>> <<<<<<<<<<< |
infC | |||
| AAAAAGCAGAAAAGCGTTTAACGCTCTTCTGCTTTTTTTGCGAGTTT >>>>>>>>>>>> <<<<<<<<<<<< |
ysdA | |||
| ATAGAGCCGCTCTCCAGCAGAGCGGCTTTTTCTATATAAAG >>>>>>>> <<<<<<<< |
rplT |
|