Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
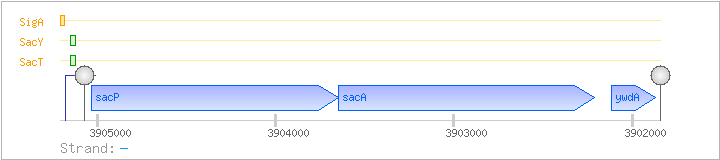
| Regulated Operon: | sacPA-ywdA |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| sacP | ipa-49d | - | 3903646..3905031 | COG1263G | scrA-BAC | |
| sacA | ipa-50d | - | 3902210..3903649 | COG1621G | ||
| ywdA | ipa-51d | - | 3901868..3902116 |
| Operon evidence: | Genome analysis |
|---|---|
| Reference: | Fouet A, et al. (1986), Debarbouille M, et al. (1990), Arnaud M, et al. (1992), Aymerich S & Steinmetz M (1992) |
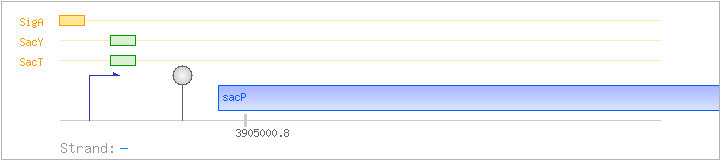
| Comments: | In-vitro transcription experiments showed that transcription terminates at the stem-loop structure upstream of sacP; transcriptional termination is alleviated in the presence of sucrose by SacT (and to a lesser degree SacY) binding to the ribonucleic antiterminator (RAT) sequence. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SacT | Positive | +21:+63 | 3905117..3905159 | ATAAGCGGGATTGTGACTGGTAAAGCAGGCAAGACCTAAAATT |
Lepesant JA, et al. (1976): Microbiology-1976, edited by David Schlessinger; pages 58-69: DB, sucrase activity Debarbouille M, et al. (1990): DB HM, sucrase activity Arnaud M, et al. (1992): SDM RG DB Arnaud M, et al. (1996): GS, in-vitro transcription |
| SacY | Positive | +21:+63 | 3905117..3905159 | ATAAGCGGGATTGTGACTGGTAAAGCAGGCAAGACCTAAAATT |
Lepesant JA, et al. (1976): Microbiology-1976, edited by David Schlessinger; pages 58-69: DB, sucrase activity Arnaud M, et al. (1996): GS, in-vitro transcription |
| SigA | Promoter | -38:+13 | 3905167..3905217 | AAAAATAGTTGACGAAAACGCTATCATGATTTATGATGAAAGCGTATTCTT |
Arnaud M, et al. (1996): RO |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| CAAGACCTAAAATTTGCGTAAATGAAAAAGGATCGCTGTGTCCTTTATTCGTTGGCGAATTTTAGGTCTTTTTT >>>>>>>>>>>>>>>>>>>>> <<<<<<<<<<<<<<<<<<<< |
sacP | |||
| ACAGGCTGCCGATGGGATCGGCAGTTTTTTCTGTGAAAA >>>>>>>> <<<<<<<< |
ywdA |
|