Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
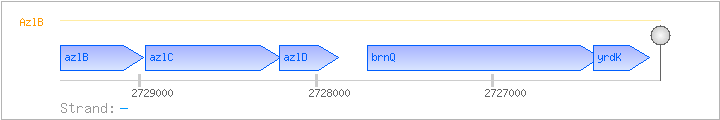
| Regulated Operon: | azlBCD-brnQ-yrdK |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
| azlB | yrdG | - | 2728979..2729452 | transcriptional regulator (Lrp/AsnC family) | COG1522 | |
| azlC | yrdH | - | 2728202..2728966 | COG1296 | ||
| azlD | yrdI | - | 2727873..2728205 | COG1687 | ||
| brnQ | yrdJ | - | 2726386..2727708 | branched-chain amino acid transporter | COG1114 | brnQ-1-BAC brnQ-BAC-1 brnQ-BAC-2 brnQ-BAC-3 brnQ-BAC-4 |
| yrdK | - | 2726111..2726428 |
| Operon evidence: | lacZ transcriptional fusions; Northern blotting (3.5 kb transcript) |
|---|---|
| Reference: | Belitsky BR, et al. (1997), Genbank U93876 |
| Comments: | This operon contains at least one internal promoter somewhere downstream from azlC (presumably in the 165 basepair gap between azlD and yrdK); Genbank U93876 shows another potential terminator in this gap. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| AzlB | Negative | ND | ND | ND |
Belitsky BR, et al. (1997): DB DP |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AAAAAGCAAACTACATACCGAAGTTTGCTTTTTTGCTCTATAC >>>>>>> <<<<<<< |
yrdK |
|