Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
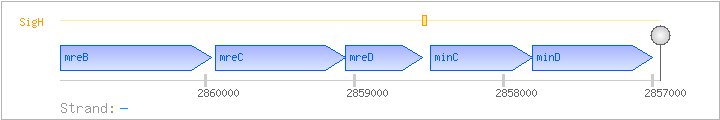
| Regulated Operon: | mreBCD-minCD |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
| mreB | divIVB | - | 2859962..2860975 | cell-shape determining protein | COG1077 | mreB-BAC mreB-CLO mreB-LAB x0825-STA |
| mreC | - | 2859059..2859931 | cell-shape determining protein | COG1792 | x0171-STA | |
| mreD | rodB | - | 2858544..2859062 | cell-shape determining protein | COG2891 | |
| minC | - | 2857811..2858491 | COG0850 | |||
| minD | divIVB1 | - | 2857003..2857809 | ATPase activator of MinC | COG2894 |
| Operon evidence: | integrative plasmids |
|---|---|
| Reference: | Varley AW & Stewart GC (1992), Lee S & Price CW (1993), Cutting S, et al. (1991) |
| Comments: | Internal promoter in front of minC. The downstream gene spoIVFA most likely belongs to a different operon. However, this was not verified experimentally. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigH | Promoter | ND | 2858506..2858547 | GTAAAAAGGATTTTATCTTTTTTTGACGAAATGAGTATGTTG |
Lee S & Price CW (1993): RG |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AAGAATCTGACAAAGCATATGCTGTGTCAGGTTTTTTTTTGTTTTT >>>>>>>>>>> <<<<<<<<<<< |
minD |
|