Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
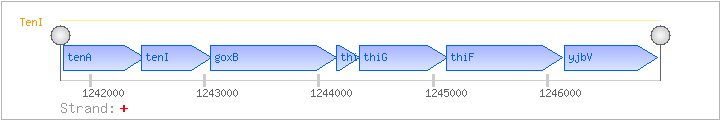
| Regulated Operon: | tenAI-goxB-thiSGF-yjbV |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
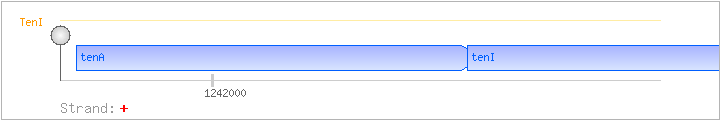
| tenA | + | 1241761..1242471 | transcriptional regulator | COG0819 | tenA-BAC x0362-STA | |
| tenI | + | 1242446..1243063 | transcriptional regulator | COG0352 | ||
| goxB | yjbR | + | 1243047..1244156 | glycine oxidase | COG0665 | |
| thiS | yjbS | + | 1244156..1244356 | COG2104 | x0478-BAC | |
| thiG | yjbT | + | 1244353..1245123 | COG2022 | ||
| thiF | yjbU | + | 1245120..1246130 | COG0476 | ||
| yjbV | + | 1246149..1246964 | COG0351 | yjbV-BAC |
| Operon evidence: | lacZ transcriptional fusions; genome analysis |
|---|---|
| Reference: | Pang AS, et al. (1991), Mironov AS, et al. (2002) |
| Comments: | The lacZ transcriptional fusions showed that tenA and tenI belong to the same operon. Genes further downstream are assigned to this operon based on genome analysis. A terminator-antiterminator structure is found upstream of tenA. Binding of thiamin pyrophosphate to the thi-box favors transcriptional termination. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| TenI | Negative | ND | ND | ND |
Pang AS, et al. (1991): DB |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| GTTATTTTACTATGCCAATTCCAAACCACTTTTCCTTGCGGGAAAGTGGTTTTTTTATTTTCAG >>>>>>>>>> <<<<<<<<<< |
tenA | |||
| TTAAATACAAGCCGATGAGATCACCAGCTGATGGTGATCTCTTTTGCGTTTGCTAC >>>>>>>>>> <<<<<<<<<< |
yjbV |
|