Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
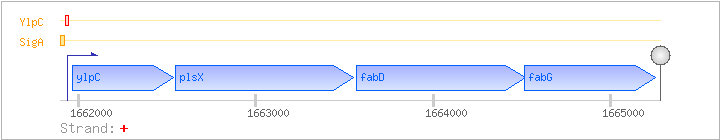
| Regulated Operon: | fapR-plsX-fabDG |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
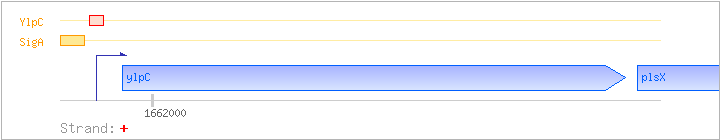
| ylpC | fapR | + | 1661967..1662533 | COG2050Q | ylpC-BAC | |
| plsX | ylpD | + | 1662547..1663548 | COG0416I | plsX-BAC | |
| fabD | ylpE | + | 1663567..1664520 | COG0331I | fabD-BAC fabD-STR | |
| fabG | ylpF | + | 1664513..1665253 | COG1028IQR | x0651-BAC |
| Operon evidence: | Genome analysis |
|---|---|
| Reference: | Schujman GE, et al. (2003) |
| Comments: |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigA | Promoter | -45:+18 | 1661892..1661954 | ATCGTTGCATTCCAATCTATGAAATTATATACTACTATTAGTACCTAGTCTTAATTGTCCGGA |
Schujman GE, et al. (2006): PE |
| YlpC | Negative | -45:+18 | 1661892..1661954 | ATCGTTGCATTCCAATCTATGAAATTATATACTACTATTAGTACCTAGTCTTAATTGTCCGGA |
Schujman GE, et al. (2003): DB RG AR GS Schujman GE, et al. (2006): FT |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| GGCGGAATGGTGATGTAAGTTTTTTCTCGAAAATTTCATCGTAGTTTCTCTAGTTTTTT >>>>>>>>>>>>>> <<<<<<<<<<<<< |
fabG |
|