Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
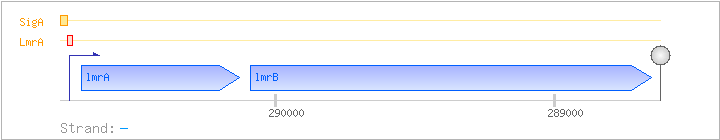
| Regulated Operon: | lmrAB |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| lmrA | yccB | - | 290132..290698 | COG1309K | ||
| lmrB | yccA | - | 288653..290092 | COG2814G |
| Operon evidence: | Northern blotting (2.1 kb transcript) |
|---|---|
| Reference: | Yoshida K, et al. (2004), Murata M, et al. (2003), Kumano M, et al. (2003) |
| Comments: | A 0.6/0.7 kb lmrA monocistronic transcript due to a readthrough terminator downstream of lmrA was also found. |
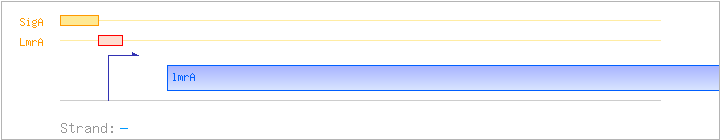
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| LmrA | Negative | -21:+21 | 290720..290761 | TTGAATCAAGATAATAGACCAGTCACTATATTTTTGATTGCA |
Murata M, et al. (2003): DB NB, promoter mutations Yoshida K, et al. (2004): GS DP FT AR, promoter mutations |
| SigA | Promoter | -48:+8 | 290733..290788 | CAATGAATTTTTCTTGACAATTGATGATTGAATCAAGATAATAGACCAGTCACTAT |
Kumano M, et al. (2003): PE |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| GAAAAGCCCCTGACTAGTGTTCAGGGGCTTTTTCATGTTTACT >>>>>>>> <<<<<<<< |
lmrB |
|