Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
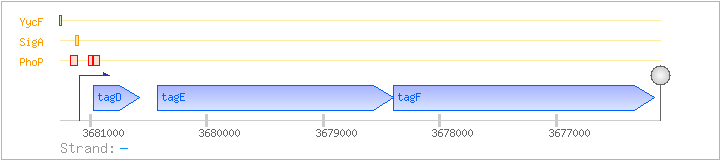
| Regulated Operon: | tagDEF |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| tagD | - | 3680581..3680970 | COG0615MI | tagD-STA | ||
| tagE | gtaD | - | 3678399..3680420 | COG0438M | ||
| tagF | rodC | - | 3676159..3678399 | COG1887M |
| Operon evidence: | Genome analysis |
|---|---|
| Reference: | Honeyman AL & Stewart GC (1989), Rosenberg M & Court D (1979) |
| Comments: | Honeyman & Stewart propose that the ATTTTATAAAAT inverted repeat in front of the terminator hairpin functions to slow the RNA polymerase prior to transcription termination. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
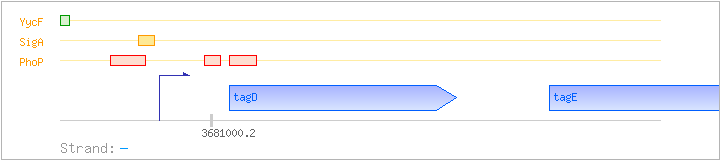
| PhoP | Negative | -81:-12 | 3681114..3681174 | GTCTGAGTGTAATTACATATTTTGGAATGTTTTTCTCCTCATGAGTACTTAACCTTTTTTA |
Liu W, et al. (1998): DB FT |
| PhoP | Negative | +122:+168 | 3680924..3680970 | ATGAAAAAAGTTATCACATATGGAACTTTTGATTTACTGCATTGGGG |
Liu W, et al. (1998): DB FT |
| PhoP | Negative | +79:+106 | 3680986..3681013 | TGTTTTTAATAGTGTTTAACATTAGGAA |
Liu W, et al. (1998): DB FT |
| SigA | Promoter | -40:+6 | 3681086..3681131 | AGTACTTAACCTTTTTTAGGCGGTATGCTATAATTAATGATCAGTG |
Mauel C, et al. (1995): PE |
| YycF | Positive | -177:-147 | 3681238..3681268 | CAAAGGATTAACATTATCTTTACATTAAATT |
Howell A, et al. (2003): HM FT |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| TGTATAGCTGTCGATTTTTCGGCAGCTTTTTAGATACTTGTT >>>>>>>>> <<<<<<<<< |
tagF |
|