Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
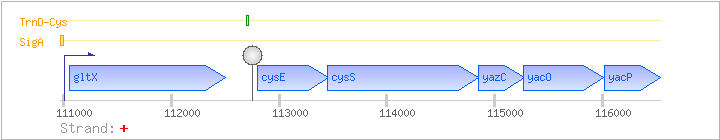
| Regulated Operon: | gltX-cysES-yazC-yacOP |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
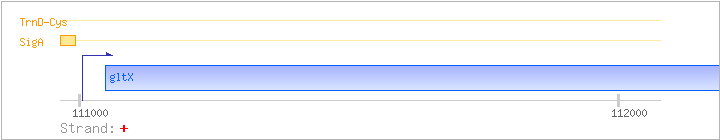
| gltX | + | 111047..112498 | COG0008J | gltX-BAC gltX-STA gltX-STR | ||
| cysE | cysA | + | 112800..113453 | COG1045E | cysE-BAC-1 cysE-BAC-2 cysE-STA | |
| cysS | snpA | + | 113450..114850 | COG0215J | ||
| yazC | + | 114851..115282 | ||||
| yacO | + | 115266..116015 | ||||
| yacP | + | 116025..116537 | COG3688R |
| Operon evidence: | Primer extension analysis of cysS; Northern blotting; S1 nuclease mapping |
|---|---|
| Reference: | Gagnon Y, et al. (1994), Pelchat M & Lapointe J (1999), BSORF |
| Comments: | Northern blotting of gltX shows a 1.7 kb transcript. The gltX-cysES-yazC-yacOP transcript is rapidly processed to generate a gltX and a cysES-yazC-yacOP mRNA molecule. RNA cleavage occurs between the last C of the stem-loop and the first T of the T-stretch of the terminator downstream of gltX. Transcriptional readthrough is expected to occur at this stem-loop if uncharged cysteine tRNA binds to the T-box motif in the nascent mRNA transcript. The tRNA anticodon binds to the UGC codon in the sequence AAGUGCGCC in the cysE leader mRNA. Northern blotting results in BSORF show a gltX, a gltX-cysES-yazC-yacOP, and a cysES-yazC-yacOP transcript. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigA | Promoter | -52:+11 | 110951..111013 | GGCTAACTTGTTTTGATGCCGGGTCTATTTGGTGGTAGAATAGATTCATACATTTTTGCCTGA |
Gagnon Y, et al. (1994): S1 |
| TrnD-Cys | Positive | ND | 112689..112721 | CTTTTCAAACAGAGTGGAACCGCGCGGTTAAAG |
Gagnon Y, et al. (1994): HM NB OV RG |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| TTAAAGCGTCTCTGTCATGTTTACATGCAGAGACGCTTTTTTTATTGGGTA >>>>>>>>>>>>>> <<<<<<<<<<<<< |
gltX |
|