Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
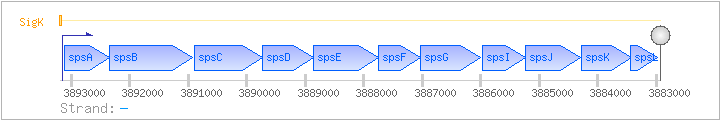
| Regulated Operon: | sps |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
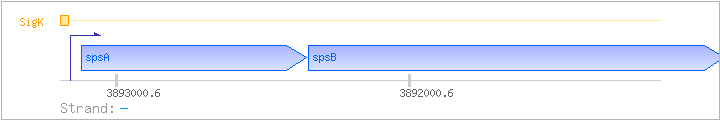
| spsA | ipa-63d | - | 3892351..3893121 | COG0463M | ||
| spsB | ipa-64d | - | 3890922..3892346 | COG1887M | spsB-STA | |
| spsC | ipa-65d | - | 3889732..3890901 | COG0399M | ||
| spsD | ipa-66d | - | 3888862..3889731 | COG0456R | ||
| spsE | ipa-67d | - | 3887741..3888862 | COG2089M | ||
| spsF | ipa-68d | - | 3887026..3887748 | COG1861M | ||
| spsG | spsH | - | 3886004..3887023 | COG3980M | ||
| spsI | ipa-71d | - | 3885239..3885979 | COG1209M | ||
| spsJ | ipa-72d | - | 3884292..3885239 | COG1088M | rfbB-BAC | |
| spsK | ipa-73d | - | 3883427..3884278 | COG1091M | ||
| spsL | ipa-73d | - | 3882979..3883434 | COG1898M |
| Operon evidence: | Genome analysis |
|---|---|
| Reference: | Glaser P, et al. (1993), Presecan E, et al. (1997) |
| Comments: |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigK | Promoter | -39:+5 | 3893155..3893198 | CCTAGCGCAACTTGAGCATAAGCAACATAAGATAACGATAGTTT |
Eichenberger P, et al. (2004): Race-PCR |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AACGTGATCAATCGAAGAGCAGAGGCATCTTCGACTTTTTTTGCCCTTTT >>>>>>>>> <<<<<<<<<< |
spsL |
|