Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
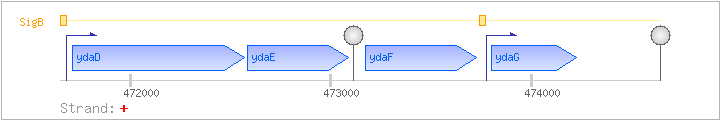
| Regulated Operon: | ydaDEFG |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
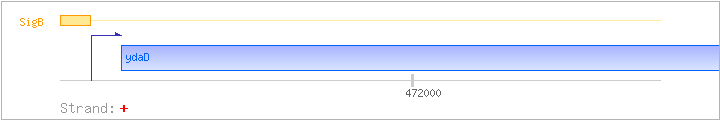
| ydaD | + | 471709..472569 | COG1028IQR | yhdF-BAC-2 yhdF-BAC-3 | ||
| ydaE | + | 472585..473088 | COG3822R | |||
| ydaF | + | 473174..473725 | COG1670J | |||
| ydaG | yzzA | + | 473803..474225 | COG3871R |
| Operon evidence: | Northern blotting (2.8 kb and 3.3 kb transcripts) |
|---|---|
| Reference: | Petersohn A, et al. (1999) |
| Comments: | The readthrough terminator downstream of ydaE leads to a 1.6 kb transcript. Both ydaDEFG transcripts terminate before reaching ydaH. An internal promoter in front of ydaG leads to monocistronic ydaG transcripts of 0.47 kb, 0.6 kb, and 0.9 kb. A 1.6 kb ydaFG transcript was also detected. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigB | Promoter | -40:+5 | 471639..471683 | AGCCATGTTTATCCAAAGAGTGTGTGAGGTACACAACAAATAGAC |
Petersohn A, et al. (1999): PE NB 2D-gel |
| SigB | Promoter | -36:+4 | 473739..473778 | TTGTTTAAATCTTCCCCGGATGTGGAAAAGTAACAGCGGA |
Petersohn A, et al. (1999): PE NB 2D-gel |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| ACAAACCGCCCGGCGTACGCCGGACGGTTTTTTTATTGCAAA >>>>>>>>> <<<<<<<<< |
ydaG | |||
| TATAAGGATGGGCTTAACAGCCCATCCTTTTTGTATTGAAAA >>>>>>>>> <<<<<<<<< |
ydaE |
|