Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
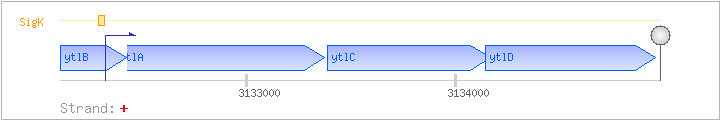
| Regulated Operon: | ytlABCD |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| ytlA | + | 3132370..3133374 | COG0715P | x0622-BAC | ||
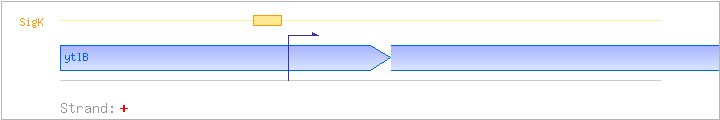
| ytlB | + | 3132105..3132422 | ||||
| ytlC | + | 3133387..3134169 | COG1116P | ytlC-BAC | ||
| ytlD | + | 3134144..3134956 | COG0600P |
| Operon evidence: | Genome analysis; downstream genes are on the opposite strand |
|---|---|
| Reference: | Genbank AF008220, Lapidus A, et al. (1997) |
| Comments: |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigK | Promoter | -36:+8 | 3132286..3132329 | GCATTCACACATTTTTATACCCATCATACGATATGTAAAGCAAA |
Eichenberger P, et al. (2004): Race-PCR |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| TGGACGAAGGGGAGGACATAAAAAACCCGGCACATGTGCCGGGTTTTTTATTCAATCC >>>>>>> <<<<<<< |
ytlD |
|