Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
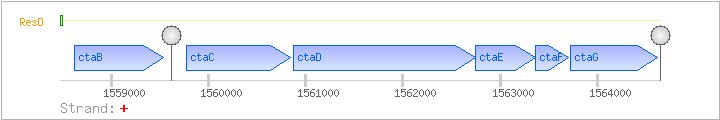
| Regulated Operon: | ctaBCDEFG |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
| ctaB | + | 1558616..1559533 | cytochrome caa3 oxydase assembly factor | COG0109 | ctaB-BAC ctaB-STA | |
| ctaC | + | 1559773..1560843 | cytochrome caa3 oxidase (subunit II) | COG1622 | ||
| ctaD | + | 1560876..1562744 | cytochrome caa3 oxidase (subunit I) | COG0843 | ctaD-BAC | |
| ctaE | + | 1562744..1563367 | cytochrome caa3 oxidase (subunit III) | COG1845 | ||
| ctaF | + | 1563370..1563702 | cytochrome caa3 oxidase (subunit IV) | COG3125 | ||
| ctaG | + | 1563729..1564622 | COG3336 |
| Operon evidence: | RNAse protection mapping of the 3' terminus of the ctaB transcript; genome analysis |
|---|---|
| Reference: | Liu X & Taber HW (1998) |
| Comments: | The RNAse protection mapping of the 3' end showed that ctaB transcription ends at the end of the stem-loop downstream of ctaB. A longer transcript was also detected, which may be due to glucose-mediated readthrough at the ctaB terminator. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| ResD | Positive | ND | 1558468..1558498 | TTGGTAAAATTCATAAAAAGTTCACAAATAA |
Liu X, et al. (1998): DB Zhang X & Hulett FM (2000): GS FT |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| CCTTGACCTCTTGTTTAATCAGGCGGCTTTACTTTTAT >>>>>>> <<<<<<< |
ctaB | |||
| ACGATATCATAAAAGGTACAGCGAAATATGCTGTACCTTTTCGTTACATTTG >>>>>>>>> <<<<<<<<< |
ctaG |
|