Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
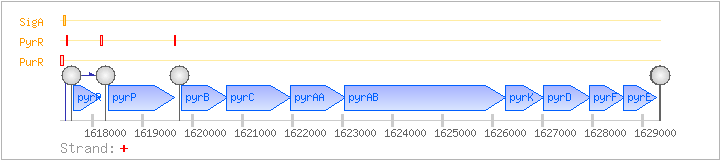
| Regulated Operon: | pyrRPBCAAABKDFE |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
| pyrR | + | 1617609..1618154 | transcriptional attenuator and uracil phosphoribosyltransferase activity (minor) | COG2065 | pyrR-BAC pyrR-LAB pyrR-STA pyrR-STR | |
| pyrP | + | 1618326..1619630 | uracil permease | COG2233 | pyrP-BAC pyrP-STA | |
| pyrB | + | 1619776..1620690 | aspartate carbamoyltransferase | COG0540 | pyrB-BAC pyrB-STA | |
| pyrC | + | 1620674..1621960 | dihydroorotase | COG0044 | pyrC-STR | |
| pyrAA | + | 1621957..1623051 | carbamoyl-phosphate synthetase (glutaminase subunit) | COG0505 | ||
| pyrAB | + | 1623036..1626251 | carbamoyl-phosphate synthetase (catalytic subunit) | COG0458 | ||
| pyrK | pyrDII, ylxD | + | 1626248..1627018 | dihydroorotate dehydrogenase (electron transfer subunit) | COG0543 | pyrK-STR |
| pyrD | pyrDI | + | 1627018..1627953 | dihydroorotate dehydrogenase (catalytic subunit) | COG0167 | pyrD-STA pyrD-STR |
| pyrF | + | 1627922..1628641 | orotidine 5'-phosphate decarboxylase | COG0284 | pyrF-STA pyrF-STR | |
| pyrE | pyrX | + | 1628620..1629270 | orotate phosphoribosyltransferase | COG0461 |
| Operon evidence: | Northern blotting (12 kb transcript) |
|---|---|
| Reference: | Turner RJ, et al. (1994), Genbank AJ000974, Genbank M59757 |
| Comments: | Northern blotting showed four transcripts (of 0.12, 0.65, 2.3, and 12 kb) corresponding to the transcriptional terminators upstream of pyrR and downstream of pyrR, pyrP, and pyrE. At these stem-loops, transcriptional termination (mediated by PyrR) occurs in the presence of excess uridine monophosphate. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
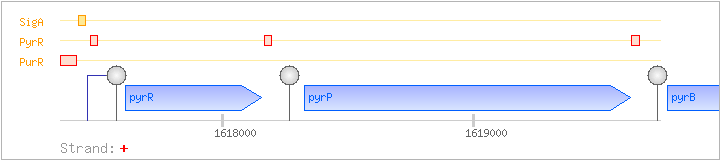
| PurR | Negative | -110:-45 | 1617349..1617414 | GGAGAATATGTCGAATTTGAAGCGCCGCTTCCCGAGGATATGGCAGAATTAATCGAAAACCTCAGA |
Shin BS, et al. (1997): GS FT |
| PyrR | Negative | ND | 1617461..1617520 | TGAATAGATTCTTTAAAACAGTCCAGAGAGGCTGAGAAGGATAACGGATAGACGGGATGC |
Lu Y, et al. (1995): DB RG Turner RJ, et al. (1994): NB RG DP OV Ghim S-Y & Switzer RL (1996): Classical genetics, RG Lu Y & Switzer RL (1996): Titration Lu Y & Switzer RL (1996): RO NB, in-vitro transcription Lu Y, et al. (1996): In-vitro transcription Bonner ER, et al. (2001): GS FT Zhang H & Switzer RL (2003): RO |
| SigA | Promoter | -40:+4 | 1617419..1617462 | ACGGTTGACAGAGGGTTTCTTTTCTGAAATAATAAACGAAGCTG |
Quinn CL, et al. (1991): NB PE |
| PyrR | Negative | ND | 1618155..1618213 | TAGATCACCTTTTTAAGCAATCCAGAGAGGTTGCAAAGAGGTGCACAACAAAGGCCCAA |
Turner RJ, et al. (1994): NB RG DP OV Lu Y, et al. (1996): DB RG Ghim S-Y & Switzer RL (1996): Classical genetics, RG Lu Y & Switzer RL (1996): Titration Lu Y & Switzer RL (1996): RO NB, in-vitro transcription Turner RJ, et al. (1998): GS DP Bonner ER, et al. (2001): GS FT SDM Zhang H & Switzer RL (2003): RO Zhang H, et al. (2005): SDM RG |
| PyrR | Negative | ND | 1619626..1619685 | TTTAAAACCTTTTAATGAAAGTCCAGAGAGGCTTGGAAGGGTTATGAAGAGAAGGAAGCT |
Lu Y, et al. (1996): DB RG Quinn CL, et al. (1991): S1 Turner RJ, et al. (1994): NB RG DP OV Ghim S-Y & Switzer RL (1996): Classical genetics, RG Lu Y & Switzer RL (1996): Titration Lu Y & Switzer RL (1996): RO NB, in-vitro transcription Bonner ER, et al. (2001): GS FT Zhang H & Switzer RL (2003): RO |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| CACCTTGTCCTAAAACCCCTCTATGCTCTGGCAGGAGGGGTTTTTTCTTCTATAT >>>>>>>>>>> <<<<<<<<<< |
pyrR | |||
| AGGTCTTTGTATGCCTCTTTGCGTAAAAAAGCAAAGAGGTTTTTTTATACAGTC >>>>>>>>> <<<<<<<<< |
pyrR | |||
| ACCATACCCCGAGTCTATCTTAGACCGGGGTTTTTTTTCAGCCTT >>>>>>>>>>> <<<<<<<<<< |
pyrP | |||
| TAAAGAGGCTTCAAAGCCTTGCTGTACTTGAAAACAGGCTGTGAGGCCTGTTTTTTTATTAATCC >>>>>>>>>>>>>> <<<<<<<<<<<<<< |
pyrE | |||
| TGAAAACAGGCTGTGAGGCCTGTTTTTTTATTAATCCT >>>>>>> <<<<<<< |
pyrE |
|