Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
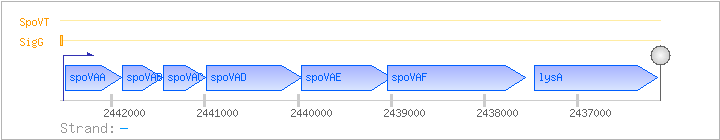
| Regulated Operon: | spoVAABCDEF-lysA |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
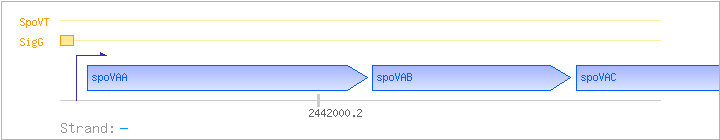
| spoVAA | - | 2441894..2442496 | ||||
| spoVAB | - | 2441459..2441884 | ||||
| spoVAC | - | 2440995..2441447 | spoVAC-BAC | |||
| spoVAD | - | 2439966..2440982 | spoVAD-BAC | |||
| spoVAE | - | 2438993..2439964 | ||||
| spoVAF | - | 2437564..2439042 | ||||
| lysA | - | 2436139..2437458 | diaminopimelate decarboxylase | COG0019 | lysA-BAC |
| Operon evidence: | integrational plasmids; Northern blotting; downstream gene is in the opposite direction |
|---|---|
| Reference: | Fort P & Errington J (1985), Azevedo V, et al. (1993), BSORF, Genbank L09228 |
| Comments: | The integrational plasmids showed that at least spoVAA through spoVAE belong to the same operon. The coding regions of spoVAE and spoVAF overlap. No terminator was found between spoVAF and lysA (Genbank L09228). Azevedo et al. found a 2.3 kb transcript orginating about 1 kb upstream of the lysA start codon, suggesting that transcription of spoVA continues into the lysA gene. However, the BSORF database contains a transcript consisting of the spoVA genes only. The lysA gene is also transcribed monocistronically as a 1.3 kb transcript. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigG | Promoter | -37:+21 | 2442500..2442557 | GATGAATGAGAACAAAATCGAACCACATACTACATATATAACCACCGAAAGATGGTGA |
Moldover B, et al. (1991): PE Sun DX, et al. (1989): DB RG |
| SpoVT | Positive | ND | ND | ND |
Bagyan I, et al. (1996): DB |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AGAAACGCCGATTTTTCGGCGCTTTTCTTATTTGAAT >>>>>> <<<<<< |
lysA |
|