Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
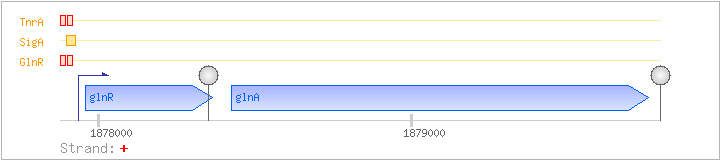
| Regulated Operon: | glnRA |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
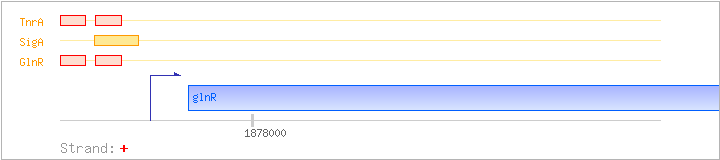
| glnR | + | 1877959..1878366 | COG0789K | glnR-STA glnR-STR | ||
| glnA | + | 1878425..1879759 | COG0174E | glnA-BAC glnA-STA glnA-STR |
| Operon evidence: | S1 nuclease mapping of the 5' and 3' end of the mRNA |
|---|---|
| Reference: | Strauch MA, et al. (1988), Fisher SH, et al. (1984) |
| Comments: | The S1 nuclease mapping of the 3' terminus showed that transcription ends in the TTTTTT stretch of the indicated terminator. The stem-loop downstream of glnR may be a readthrough terminator, or alternatively a transcriptional pause site or RNA processing site. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| GlnR | Negative | -58:-42 | 1877877..1877893 | TGTTAAGAATCCTTACA |
Gutowski JC, et al. (1992): DB FT |
| GlnR | Negative | -35:-19 | 1877900..1877916 | TGACACATAATATAACA |
Gutowski JC, et al. (1992): DB FT |
| SigA | Promoter | -49:+17 | 1877886..1877951 | TCCTTACATCGTATTGACACATAATATAACATCACCTATAATGAAACTAAGTTAAGAAAAGGAGGA |
Strauch MA, et al. (1988): RO |
| TnrA | Negative | -58:-42 | 1877877..1877893 | TGTTAAGAATCCTTACA |
Wray LV Jr, et al. (1996): DB |
| TnrA | Negative | -35:-19 | 1877900..1877916 | TGACACATAATATAACA |
Wray LV Jr, et al. (1996): DB |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| CTCAATCCCTTGGCACTAAAAGTGTCAGGGGATTTTTTATGTTAATA >>>>>>>>>>>> <<<<<<<<<<<< |
glnA | |||
| AGGGAATACATTCCGTCAAGGCGACATGTCCCGCTTCTTTCATTAATCC >>>>>>>>>>>>>>>> <<<<<<<<<< |
glnR |
|