Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
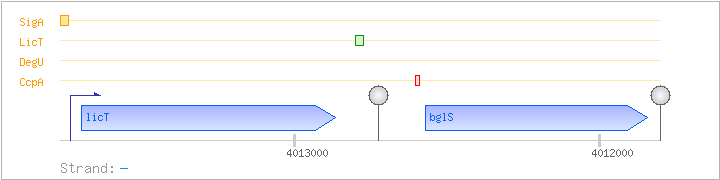
| Regulated Operon: | licT-bglS |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| licT | - | 4012866..4013699 | COG3711K | x0231-BAC | ||
| bglS | licS | - | 4011842..4012570 | COG2273G |
| Operon evidence: | Northern blotting (2.4 kb transcript) |
|---|---|
| Reference: | Schnetz K, et al. (1996) |
| Comments: | The terminator structure downstream of licT leads to a 0.9 kb transcript. In the presence of the beta-glucan lichenan, LicT binds to the ribonuclic antiterminator (RAT) sequence just upstream, alleviating termination at this stem-loop. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| CcpA | Negative | ND | 4012590..4012603 | AGAAAACGCTTTCA |
Kruger S, et al. (1993): SDM DB |
| DegU | Positive | ND | ND | ND |
Aymerich S et al. (1986): DB, glucanase activity |
| LicT | Positive | ND | 4012766..4012805 | TGGTAGGATTGTTACTGATAAAGCAGGCAAAACCTAAATT |
Steinmetz M & Aymerich S (1986): HM Schnetz K, et al. (1996): beta-glucanase activity, RG S1 NB DB OV |
| SigA | Promoter | -48:+5 | 4013730..4013782 | TTTCCCTGATGATATTGACCTAGTACAAAAAGAATGTACAATCAAGATATAGT |
Schnetz K, et al. (1996): PE |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| TGAAAGAGCCTGCTGCAATATAGCAGGCTCTTATGATTGTAATGA >>>>>>>>>> <<<<<<<<<< |
bglS | |||
| TAAATTGCAATGAGTGCGGATCATCTCTGTCTGTGCTGATGGTAATTTAGGTTTTTATTT >>>>>>>>>>>>>>>>> <<<<<<<<<<<<<<<<< |
licT |
|