Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2008
Contact: Kenta Nakai |
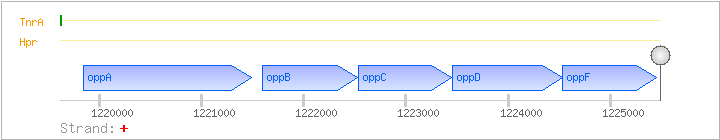
| Regulated Operon: | oppABCDF |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Conserved groups |
|---|---|---|---|---|---|---|
| oppA | spo0KA | + | 1219849..1221486 | COG4166E | oppA-BAC-1 oppA-BAC-2 oppA-LAB x0571-BAC x0772-BAC x0799-BAC x1530-STA | |
| oppB | spo0KB | + | 1221594..1222529 | COG0601EP | oppB-BAC oppB-STA stu1441-STR x0140-BAC x0484-BAC x1218-BAC x1303-BAC | |
| oppC | spo0KC | + | 1222533..1223450 | COG1173EP | x0178-BAC x0494-BAC x1458-BAC | |
| oppD | spo0KD | + | 1223455..1224531 | COG0444EP | appD-BAC-1 appD-STA | |
| oppF | spo0KE | + | 1224533..1225450 | COG1123R |
| Operon evidence: | Northern blotting (6.0 kb transcript) |
|---|---|
| Reference: | Perego M, et al. (1991), Yoshida K, et al. (2003) |
| Comments: | The secondary structure of the RNA corresponding to the oppA-oppB intergenic region may enhance the stability of the mRNA molecule (Perego), or function as a readthrough terminator (1.8 kb oppA transcript found by Yoshida). |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
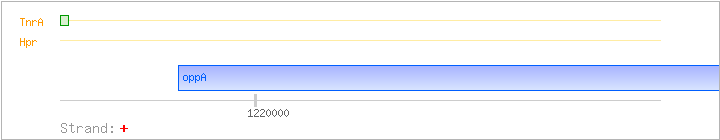
| Hpr | Negative | ND | ND | ND |
Koide A, et al. (1999): DB OV |
| TnrA | Positive | ND | 1219618..1219634 | TGGAAGAAAAACTAACG |
Yoshida K, et al. (2003): AR HM GS |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| ATCAATCCTTCAAGAGATTTCTCTTGAAGGATTTTTTTGCGTCTTC >>>>>>>>>>> <<<<<<<<<<< |
oppF |
|