Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
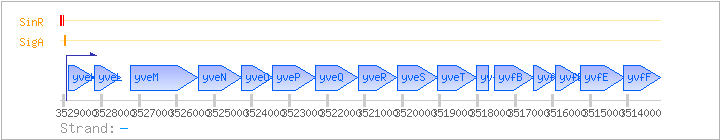
| Regulated Operon: | eps |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
| yveK | epsA | - | 3528181..3528885 | COG3944 | ||
| yveL | epsB | - | 3527492..3528175 | COG0489 | ||
| yveM | epsC | - | 3525437..3527233 | COG1086 | ||
| yveN | epsD | - | 3524280..3525425 | COG0438 | ||
| yveO | epsE | - | 3523447..3524283 | COG0463 | ||
| yveP | epsF | - | 3522300..3523454 | COG0438 | ||
| yveQ | epsG | - | 3521200..3522303 | |||
| yveR | epsH | - | 3520141..3521175 | COG0463 | ||
| yveS | epsI | - | 3519060..3520136 | COG5039 | ||
| yveT | epsJ | - | 3518029..3519063 | COG0463 | ||
| yvfA | epsK | - | 3517703..3518032 | |||
| yvfB | epsK | - | 3516516..3517553 | |||
| yvfC | epsL | - | 3515911..3516519 | COG2148 | yvfC-LAC | |
| yvfD | epsM | - | 3515264..3515914 | COG0110 | yvjF-LAC | |
| yvfE | epsN | - | 3514093..3515259 | |||
| yvfF | epsO | - | 3513146..3514114 | COG5039 |
| Operon evidence: | Genome analysis. Transcriptional fusions of lacZ to the yveK and yveM upstream regions indicate that yveM does not have its own promoter (D. Kearns, personal communication). However, Northern blotting experiments by Yoshida show a 1.2 kb yveKL transcript, which may be due to a readthrough terminator downstream of yveL. Note that yvfA and yvfB constitute one gene (epsK); this gene is listed as two ORFs due to a sequencing error (Kearns et al). |
|---|---|
| Reference: | Yoshida K, et al. (2003), Kearns DB, et al. (2005) |
| Comments: |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigA | Promoter | -43:+6 | 3528936..3528985 | TTTTGCAATTTTTAAATAATAACGTTTTCTTTTATAATCCAATCATTAAC |
Kearns DB, et al. (2005): PE |
| SinR | Negative | -163:-124 | 3529066..3529105 | TCGTTATTTCGTTCATTATAAGGAATTTTTCGTTCTTTAT |
Kearns DB, et al. (2005): GS FT RG |
| SinR | Negative | -73:-48 | 3528990..3529015 | TTTGTTCTCTAAAGAGAACTTATTGG |
Kearns DB, et al. (2005): GS FT RG |

|