Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
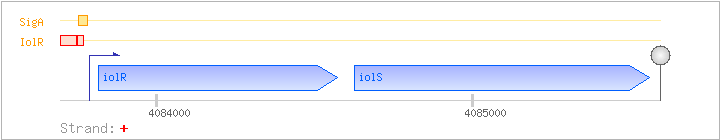
| Regulated Operon: | iolRS |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
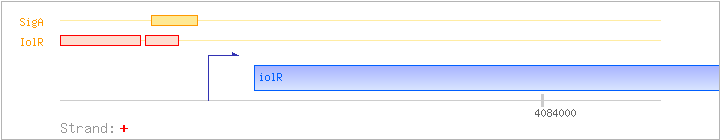
| iolR | + | 4083818..4084573 | transcriptional regulator (DeoR family) | COG1349 | ||
| iolS | yxbF | + | 4084627..4085559 | COG0667 |
| Operon evidence: | Northern blotting (2.0 kb transcript); S1 nuclease mapping of the 3' end (transcription ends in the CTGT region just in front of the T-stretch) |
|---|---|
| Reference: | Yoshida KI, et al. (1997) |
| Comments: |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| IolR | Negative | -94:-43 | 4083695..4083746 | AGAAGGCCGTCCAATCTTGCAAAAGATATACACCGATGTTATCATTTTTAAG |
Yoshida KI, et al. (1997): DB DP HB |
| IolR | Negative | -40:-19 | 4083749..4083770 | ACTATTGATTAACTTTTGGTTT |
Yoshida KI, et al. (1999): GS FT |
| SigA | Promoter | -39:+5 | 4083750..4083793 | CTATTGATTAACTTTTGGTTTTTATTATATATTTATGTTACGTA |
Yoshida KI, et al. (1997): PE |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| GAAAACAGCCTTCTCCAATGGAGAAGGCTGTTTTTTTGTGCGATA >>>>>>>>>>> <<<<<<<<<<< |
iolS |
|