Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
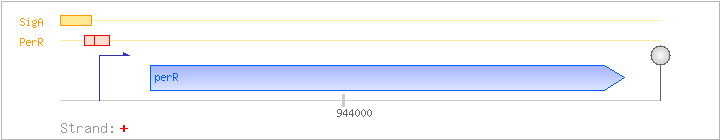
| Regulated Operon: | perR |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
| perR | ygaG | + | 943822..944259 | transcriptional regulator (Fur family) | COG0735 | x0088-BAC |
| Operon evidence: | downstream gene is on the opposite strand |
|---|---|
| Reference: | Fuangthong M, et al. (2002) |
| Comments: |
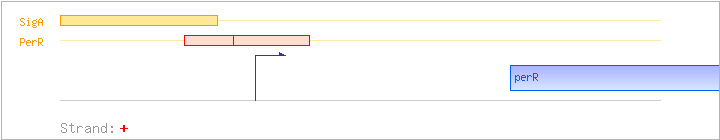
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| PerR | Negative | -13:+13 | 943762..943787 | TTACACTAATTATAAACATTACAATG |
Fuangthong M, et al. (2002): FT |
| PerR | Negative | -4:+18 | 943771..943793 | TTATAAACATTACAATGTAAGAA |
Fuangthong M, et al. (2002): FT |
| SigA | Promoter | -41:+5 | 943734..943779 | ATCAATTGACAAATTATCGTAGAAAGAGTTACACTAATTATAAACA |
Fuangthong M, et al. (2002): PE HM |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AAAAATAAGCTGACCGCACGAAACGGTCAGCTTATTTTATTTCGACATT >>>>>>>>>>> <<<<<<<<<<< |
perR |
|