Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
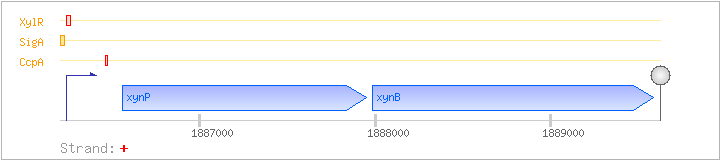
| Regulated Operon: | xynPB |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
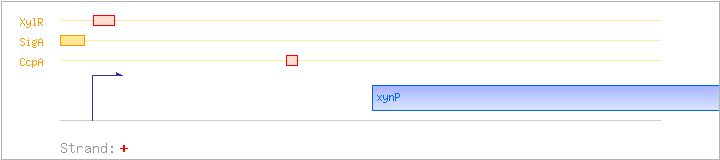
| xynP | ynaJ, xynC | + | 1886560..1887951 | H+-symporter | COG2211 | |
| xynB | ynaK | + | 1887982..1889583 | xylan beta-1,4-xylosidase | COG3507 |
| Operon evidence: | Genome analysis; downstream gene is on the opposite strand |
|---|---|
| Reference: | Hastrup S: Genetics and Biotechnology of Bacilli Vol. 2: pp. 79-83 (1988) |
| Comments: |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| CcpA | Negative | +219:+242 | 1886457..1886480 | TGTTTTGAAAGCGCTTTTATAAAA |
Lindner C, et al. (1994): DB HB HM RG Galinier A, et al. (1999): DP RG FT |
| SigA | Promoter | -47:+11 | 1886192..1886249 | CTGAAAAAGATGTTGAAAAAGTCGAAAGGATTTTATAATATTAAGTCAAGTTAGTTTG |
Hastrup S (1988): Genetics and biotechnology of Bacilli, vol.2 p.79-83: S1 Galinier A, et al. (1999): PE DP |
| XylR | Negative | -3:+35 | 1886236..1886273 | GTCAAGTTAGTTTGTTTGATCAACAAACTAATGAAATG |
Hastrup S (1988): Genetics and biotechnology of Bacilli, vol.2 p.79-83: ND Kreuzer P, et al. (1989): HM Galinier A, et al. (1999): RG |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| TTGCACATGAAAAAGGAGATTTCTATTTTAGAACTCCTTTTTCATATGAGAA >>>>>>>>>>> <<<<<<<<< |
xynB |
|