Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
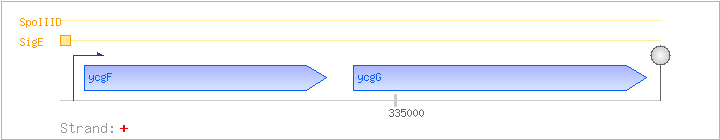
| Regulated Operon: | ycgFG |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
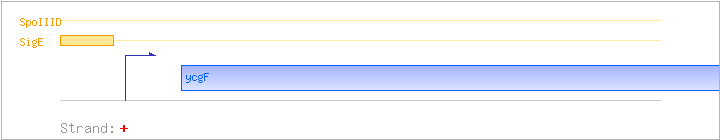
| ycgF | + | 334192..334821 | COG1280 | |||
| ycgG | + | 334891..335652 | COG3403 |
| Operon evidence: | downstream gene ycgH is in the opposite direction; no experimental evidence that ycgF and ycgG are in the same operon |
|---|---|
| Reference: | Eichenberger P, et al. (2003) |
| Comments: | Northern blotting results in BSORF show a ycgEFG and a ycgG transcript. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| SigE | Promoter | -37:+9 | 334125..334170 | TTGTGCATAGCTTGGCCCGTTCCCGAATAAATTGTACAAGTTACAT |
Eichenberger P, et al. (2003): AR, 5'-RACE-PCR |
| SpoIIID | Negative | ND | ND | ND |
Eichenberger P, et al. (2004): GS AR |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| CTGGCAGGGCGATCTTTGTGACCCTACTTTTTTTGATAGATC >>>>>> <<<<<<< |
ycgG |
|