Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
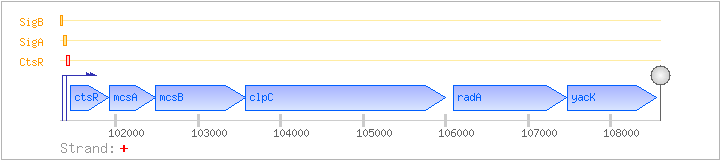
| Regulated Operon: | ctsR-mcsAB-clpC-radA-yacK |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
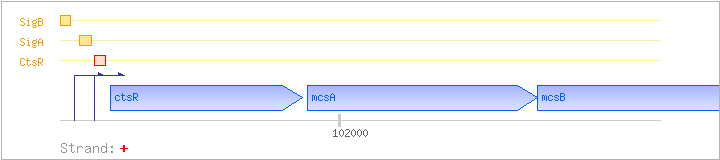
| ctsR | yacG | + | 101446..101910 | transcriptional regulator | COG4463 | ctsR-BAC ctsR-STA ctsR-STR |
| mcsA | yacH | + | 101924..102481 | COG3880 | ||
| mcsB | yacI | + | 102481..103572 | COG3869 | ||
| clpC | mecB | + | 103569..106001 | class III stress response-related ATPase | COG0542 | clpC-BAC clpC-STA |
| radA | sms, yacJ | + | 106093..107469 | DNA repair protein homolog | COG1066 | radA-BAC |
| yacK | + | 107473..108555 | COG1623 |
| Operon evidence: | Northern blotting (7.0 kb transcript) and S1 nuclease mapping of the 3' end |
|---|---|
| Reference: | Kruger E, et al. (1996) |
| Comments: | A ctsR-mcsAB-clpC transcript and various transcripts in the radA-yacKLMN region are listed in BSORF. The 3' nuclease mapping showed two weak termination signals in the AT-rich region at the end of a proposed terminator structure eight base pairs downstream of the stop codon (i.e., in the TTAAAA region in the left arm of the indicated stem-loop). However, mfold cannot detect such a stem-loop structure. Northern blotting results in BSORF show a ctsR-mcsAB-clpC transcript. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| CtsR | Negative | +1:+29 | 101406..101434 | AAAGTCAAATATAGTCAAAGTCAGTAAAG |
Derre I, et al. (1999): GS FT |
| SigA | Promoter | -43:+9 | 101363..101414 | CGAGAAAGTTGAAAATTCTCGAGAAACGGCTTATAGTAAGATTAAAGTCAAA |
Kruger E, et al. (1996): NB PE |
| SigB | Promoter | -46:+6 | 101311..101362 | ATCAGCAATCAGGTTTTGTGGACCGGGAAAATGGAAATAATGAAGGATAGAG |
Kruger E, et al. (1996): NB PE |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| TAAAACCTTATGAATACGGGTATATTAATGTTGGTTTTTGTTTATTCTG >>>>>>>>>>> <<<<<<<<<<<< |
yacK |
|