Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
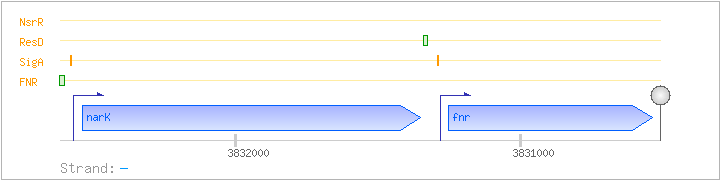
| Regulated Operon: | narK-fnr |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
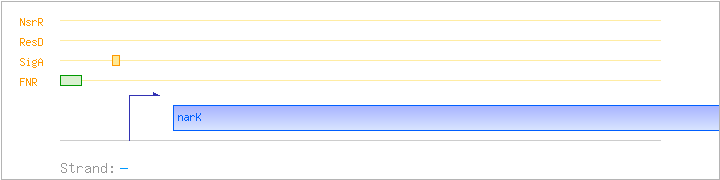
| narK | - | 3831350..3832537 | nitrite extrusion protein | COG2223 | narK-BAC | |
| fnr | - | 3830535..3831251 | transcriptional regulator (FNR/CRP family) | COG0664 | x0456-BAC |
| Operon evidence: | Northern blotting (2.0 kb transcript) |
|---|---|
| Reference: | Cruz Ramos H, et al. (1995) |
| Comments: | Internal promoter in front of fnr, leading to a 0.7 kb monocistronic fnr transcript. A stem-loop structure between narK and fnr apparently does not function as a terminator |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| FNR | Positive | -52:-31 | 3832599..3832620 | ACGTGTGATGTAATTCACAATC |
Cruz Ramos H, et al. (1995): DB PE Reents H, et al. (2006): AR |
| SigA | Promoter | -52:+8 | 3832561..3832620 | ACGTGTGATGTAATTCACAATCCTGTTTGGCTAGTTTTTGTATGATAAGACTGATTATTG |
Cruz Ramos H, et al. (1995): PE |
| ResD | Positive | -62:-44 | 3831324..3831341 | CATTCACAAGATTGTTAG |
Nakano MM, et al. (1996): DB RG Nakano MM, et al. (2001): RG |
| SigA | Promoter | -48:+18 | 3831262..3831327 | TTAGTTTTTTCTCGATGGGTACAAATCAATTCACGTTACAATAAACACACAACGTGACTTTCTTCG |
Cruz Ramos H, et al. (1995): PE |
| NsrR | Negative | ND | ND | ND |
Nakano MM, et al. (2006): RG |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| AAAAACAAGGACAGCGGTGTCCTTGTTTTTTTACCTTGTG >>>>>>>> <<<<<<<< |
fnr |
|