Copyright: Human Genome Center, Inst. Med. Sci., Univ. Tokyo; 1999-2006
Contact: Kenta Nakai |
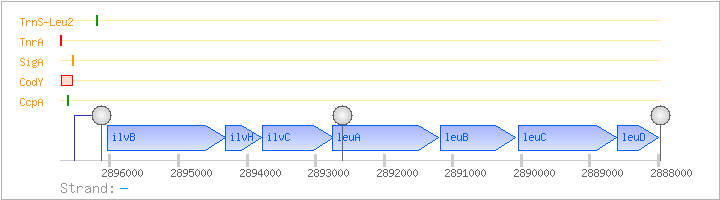
| Regulated Operon: | ilvBHC-leuABCD |
| Genes | Synonyms | Direction | Genome position | Function | COG ID | Genus clusters |
|---|---|---|---|---|---|---|
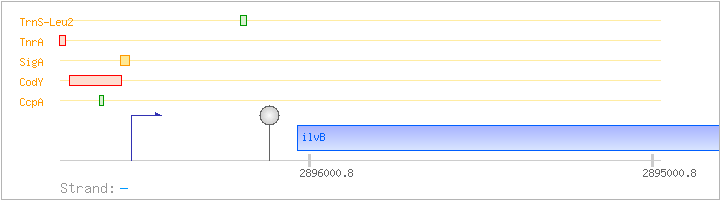
| ilvB | - | 2894312..2896036 | acetolactate synthase (acetohydroxy-acid synthase) (large subunit) | COG0028 | ilvB-2-BAC ilvB-BAC ilvB-STA | |
| ilvH | ilvN | - | 2893791..2894315 | acetolactate synthase (acetohydroxy-acid synthase) (small subunit) | COG0440 | |
| ilvC | - | 2892746..2893774 | ketol-acid reductoisomerase (acetohydroxy-acid isomeroreductase) | COG0059 | ilvC-BAC ilvC-STA ilvC-STR | |
| leuA | - | 2891203..2892759 | 2-isopropylmalate synthase | COG0119 | leuA-STA | |
| leuB | leuC | - | 2890085..2891182 | 3-isopropylmalate dehydrogenase | COG0473 | leuB-BAC leuC-STA |
| leuC | - | 2888617..2890035 | 3-isopropylmalate dehydratase (large subunit) | COG0065 | leuC-STA | |
| leuD | - | 2888005..2888604 | 3-isopropylmalate dehydratase (small subunit) | COG0066 |
| Operon evidence: | Northern blotting (8.5 kb transcript) |
|---|---|
| Reference: | Grandoni JA, et al. (1992), Grandoni JA, et al. (1993), Mader U, et al. (2004) |
| Comments: | RNA processing leads to a 5.8 kb ilvC-leuABCD transcript. Runoff transcription, primer extension, promoter deletions, reporter gene fusions, and site-directed mutagenesis showed that readthrough occurs at the terminator in the ilvB promoter when uncharged tRNA-Leu binds to the T-box sequence motif in the nascent tRNA transcript. The tRNA anticodon binds to the CUC codon in the sequence GAACUCGCC in the ilvB leader mRNA. The stem-loop structure in the 5' region of leuA may be a readthrough terminator or a mRNA cleavage site, leading to a monocistronic 1.2 kb ilvC transcript. The second stem-loop shown downstream of leuA is an alternative secondary structure in this region that is followed by a T-stretch, suggesting a readthrough terminator. |
| Binding factor |
Regulation | Location | Absolute position | Binding seq.(cis-element) | Experimental evidence |
|---|---|---|---|---|---|
| CcpA | Positive | -106:-53 | 2896571..2896624 | TGTACCAATAATGAAAGCGTATACAATATAGATTGATTAATCAAAATTGTCTAA |
Tojo S, et al. (2005): FT Shivers RP, et al. (2005): FT |
| CodY | Negative | -199:+1 | 2896518..2896717 | TCTTCCATGTTTATATAAAATTAAATAATTCTGGTGTATTCTTGATAGATTAATAAAAAAATTATAAAAACTTTTAACATTTGCAATTCCTTTTGTACCAATAATGAAAGCGTATACAATATAGATTGATTAATCAAAATTGTCTAATAATTTTAAAAAATGCTGTTGACACTGCGTCCAAAGCGGCGTAATATGAGTTC |
Molle V, et al. (2003): AR RG, ChIP-on-chip, GS Mader U, et al. (2004): NB Shivers RP, et al. (2004): FT |
| SigA | Promoter | -45:+19 | 2896500..2896563 | AAAAAATGCTGTTGACACTGCGTCCAAAGCGGCGTAATATGAGTTCAACAAAAGATAAATGCAA |
Grandoni JA, et al. (1992): PE SDM RG |
| TnrA | Negative | -210:-193 | 2896696..2896745 | CATATACGAACCATATCGTGTTATAAAATCTTCCATGTTTATATAAAATT |
Tojo S, et al. (2004): FT |
| TrnS-Leu2 | Positive | ND | 2896184..2896202 | AAACAAGGGTGGTACCGCG |
Grandoni JA, et al. (1993): DP RG PE SDM, in-vitro transcription Garrity DB & Zahler SA (1994): DB RG |
| Terminator sequence | Absolute position | Position from stop codon | Free energy [kcal/mol] |
Downstream of |
|---|---|---|---|---|
| TTTTCGCCCCTTTTAGCTATCGCAGTTACTGCGCGGCTGATTGTGGGCGGAAGGGCTTTTTTTATTGAATA >>>>>>>>>>>>>>>>>>>>>>> <<<<<<<<<<<<<<<<<<<<< |
ilvB | |||
| CATCATTGAAGCGGGATTTCCCGCTTCGTCCCGAGGTGACTTTTTAGCTGTTCAGGA >>>>>>>>> <<<<<<<<< |
leuA | |||
| TTTCCCGCTTCGTCCCGAGGTGACTTTTTAGCTGTTCAG >>>>>>> <<<<<<< |
leuA | |||
| AAAAAGCGGCCGGTATTTGTCCGGCGGCTTTTTTTGCCTGGTG >>>>>>>>> <<<<<<<<<< |
leuD |
|